Generate weight matrix for focal operations using map units
Source:R/fastfocal_weights.R
fastfocal_weights.RdBuilds an unnormalized (or normalized) kernel from map units. Circle uses a center-distance rule (include if center <= d). Gaussian interprets d as sigma in map units and truncates at 3 sigma, matching terra::focalMat(..., type = "Gauss").
Arguments
- x
SpatRaster (used for resolution; assumes square pixels).
- d
numeric. Radius in map units for most kernels; sigma in map units for "gaussian"/"Gauss".
- w
character. One of: "rectangle","circle","circular","gaussian","Gauss","pareto","idw", "exponential","triangular","cosine","logistic","cauchy","quartic","epanechnikov".
- normalize
logical. If TRUE (default), scale weights to sum to 1.
- plot
logical. If TRUE, plots the kernel.
Examples
# Small raster (resolution = 1 map unit)
r <- terra::rast(nrows = 5, ncols = 5, xmin = 0, xmax = 5, ymin = 0, ymax = 5)
# Circle: d is a radius in map units -> here cell_radius = 2 -> 5x5 kernel
Kc <- fastfocal_weights(r, d = 2, w = "circle", normalize = TRUE)
dim(Kc) # 5 x 5
#> [1] 5 5
round(sum(Kc), 6) # ~1
#> [1] 1
# Gaussian: d is sigma in map units, truncated at 3 sigmas
Kg <- fastfocal_weights(r, d = 1, w = "gaussian", normalize = TRUE)
dim(Kg) # 7 x 7 (since 2*ceil(3*sigma) + 1)
#> [1] 7 7
round(sum(Kg), 6) # ~1
#> [1] 1
# \donttest{
# Quick visualization (kept out of CRAN's main run)
fastfocal_weights(r, d = 2, w = "circle", normalize = TRUE, plot = TRUE)
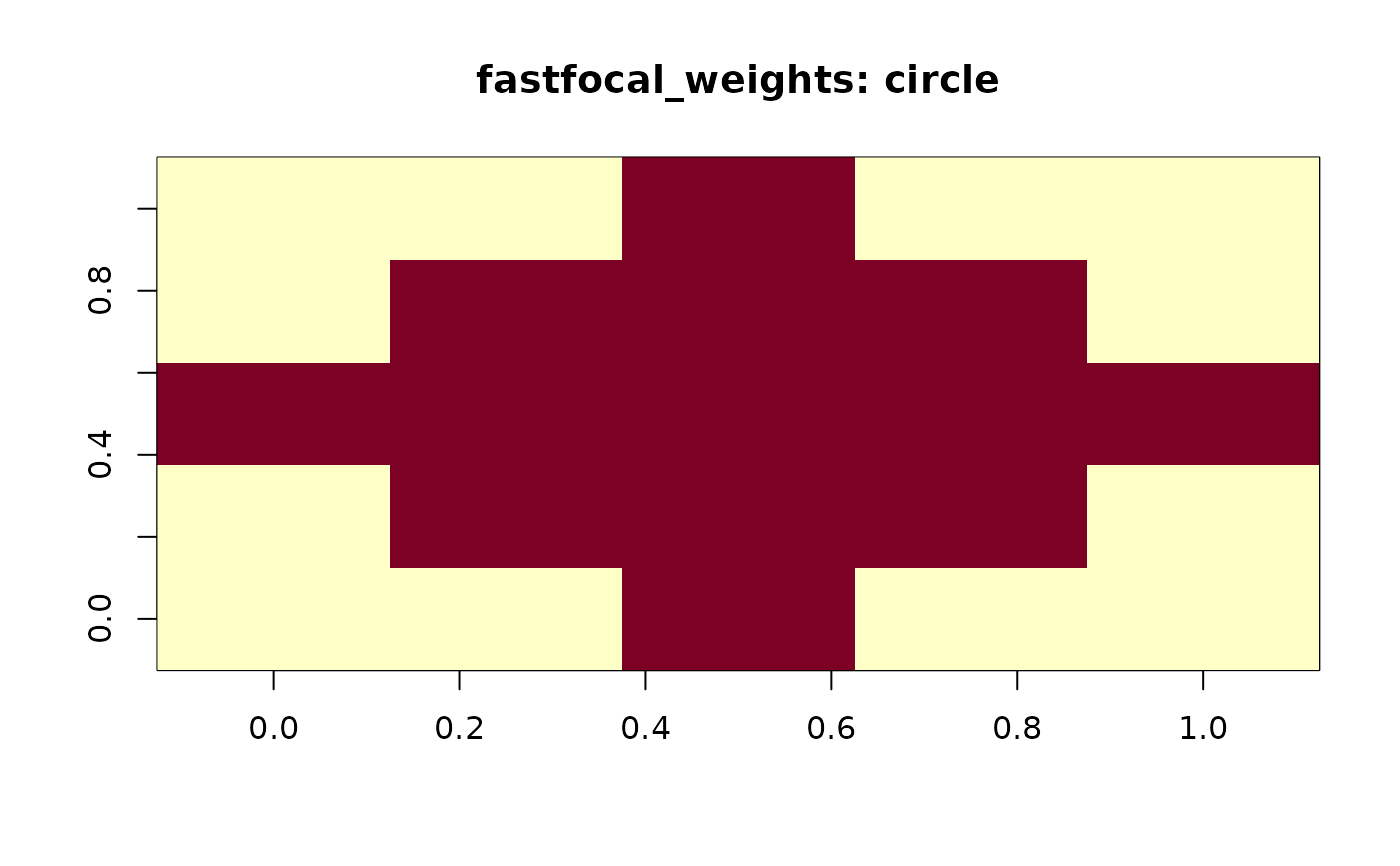 #> [,1] [,2] [,3] [,4] [,5]
#> [1,] 0.00000000 0.00000000 0.07692308 0.00000000 0.00000000
#> [2,] 0.00000000 0.07692308 0.07692308 0.07692308 0.00000000
#> [3,] 0.07692308 0.07692308 0.07692308 0.07692308 0.07692308
#> [4,] 0.00000000 0.07692308 0.07692308 0.07692308 0.00000000
#> [5,] 0.00000000 0.00000000 0.07692308 0.00000000 0.00000000
# }
#> [,1] [,2] [,3] [,4] [,5]
#> [1,] 0.00000000 0.00000000 0.07692308 0.00000000 0.00000000
#> [2,] 0.00000000 0.07692308 0.07692308 0.07692308 0.00000000
#> [3,] 0.07692308 0.07692308 0.07692308 0.07692308 0.07692308
#> [4,] 0.00000000 0.07692308 0.07692308 0.07692308 0.00000000
#> [5,] 0.00000000 0.00000000 0.07692308 0.00000000 0.00000000
# }